Projects
-
Subject-Specific Analysis of Anatomical Shape Changes
-
Explainable Imaging Biomarker Extraction
-
Representative Image and Spatio-Temporal Trajectory Estimation
-
Full 3D Medical Image Synthesis using Deep Generative Model
Subject-Specific Analysis of the Anatomical Shape Change
Morphological changes of anatomical structures are widely studied as potential biomarkers that allow researchers and clinicians to track disease progression, even before clinical diagnosis. In this project, we proposed novel longitudinal modeling methods that can analyze the subject-specific associations of the morphological changes and premanifest disease progression on Riemannian manifold. Applications to real-world data from a large clinical study for a Huntington’s disease, a neurodegenertative disease without a known therapy, demonstrate the ability and feasibility of the proposed methods for identification of potential biomarkers at the prodromal stage before clinical diagnosis. The methods that were proposed in this project include both cross-sectional and longitudinal regression models, which are generic approaches with potentials to be applied to a wide variety of medical image analysis applications with manifold-valued data.



Explainable Imaging Biomarker Extraction
There are numerous anatomical characteristics that can be observed in medical images. Because those anatomical characteristics are entangled together in a medical image, it is challenging to analyze the association of an individual and explainable imaging characteristic and a disease. This project aims to provide a novel tool leveraging deep learning models that decomposes medical images to a set of explainable imaging features (e.g., expansion/atrophy of the ventricles and white matter hyperintensity progression for stroke patients) that can be used as essential biomarkers of disease severity or disease progression. This project is currently ongoing with a close collaboration with MIT CSAIL Medical Vision Group.
Geometric Change
Entangled Image Change
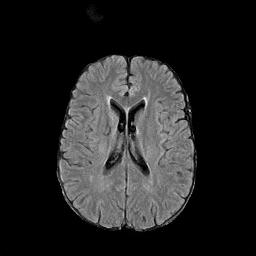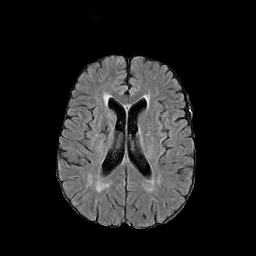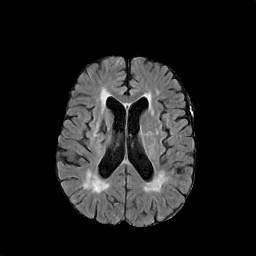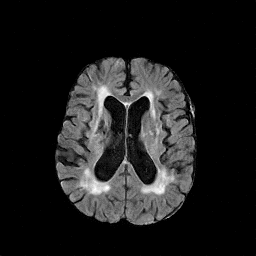
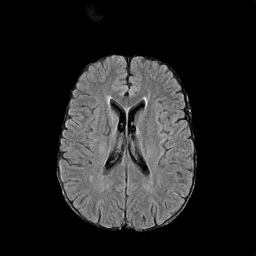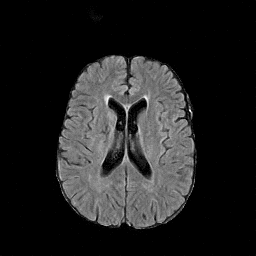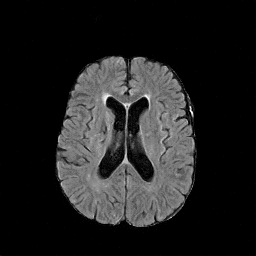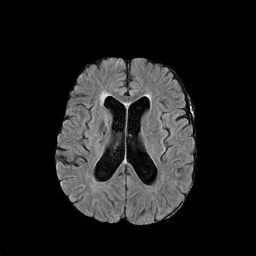
Non-Geometric Change
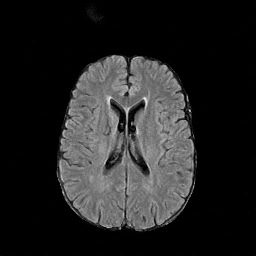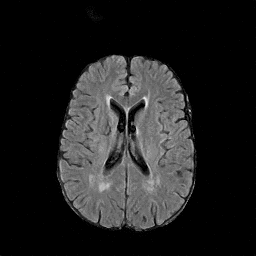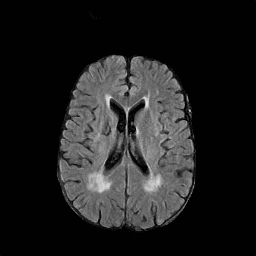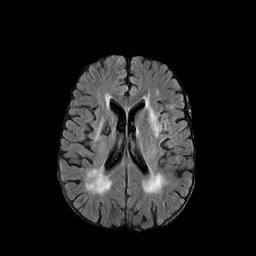
Representative Image and Spatio-Temporal Trajectory
The anatomical characteristics and changes thereof regarding clinical factors (e.g., disease progression or aging) that are shared by a group of multiple subjects is of utmost interest in medical research. Conventionally in medical image analysis, the estimation of the representative image and image trajectory has been done by geometric (conditional) atlas estimation. However, the geometric atlas estimation inherently does not account non-geometric anatomical characteristics that are essential for many clinical applications (e.g., acute stroke lesions or traumatic brain injuries). In this work, we aim to provide novel methods to estimate such representative images while accounting for those non-geometric characteristics by leveraging the state-of-the-art deep generative model and its latent space operations. This work is in preparation for a journal submission with close collaborations with MIT CSAIL Medical Vision Group and A.A. Martinos Center at MGH.


Full 3D Medical Image Synthesis using Deep Generative Model
Image synthesis via Generative Adversarial Networks (GANs) of three-dimensional (3D) medical images has great potential that can be extended to many medical applications, such as, image enhancement and disease progression modeling. Current GAN technologies for 3D medical image synthesis must be significantly improved to be suitable for real-world medical problems. In this project, we extend the state-of-the-art StyleGAN2 model, which natively works with two-dimensional images, to enable 3D image synthesis. In addition to the image synthesis, we investigate the behavior and interpretability of the 3D-StyleGAN via style vectors inherited form the original StyleGAN2 that are highly suitable for medical applications: (i) the latent space projection and reconstruction of unseen real images, and (ii) style mixing. The model can be applied to any 3D volumetric images. We demonstrate the 3D-StyleGAN’s performance and feasibility with∼12,000 three-dimensional full brain MRT1 images. Furthermore, we explore different configurations of hyper-parameters to investigate potential improvement of the image synthesis with larger networks. The codes and pre-trained networks are available online https://github.com/sh4174/3DStyleGAN
